Outputs
Pandora will store several data in the output folder, the tree structure is defined in the file pandora/output_tree_design.py.
Saved images
left_disparity.tif, right_disparity.tif : disparity maps in left and right image geometry.
left_validity_mask.tif, right_validity_mask.tif : the Validity mask in left image geometry, and right. Note that bits 4, 5, 8 and 9 can only be calculated if a validation step is set.
left_confidence_measure.tif, right_confidence_measure.tif : multi-band image, each band represents a confidence measurement, depending on what is activated.
Standard deviation of pixel intensity inside matching cost window. see Cost volume confidence. This band is named confidence_from_intensity_std.
Ambiguity measurement, see Cost volume confidence. This band is named confidence_from_ambiguity.
Mininum and maximum risk measurement, see Cost volume confidence. Those bands are named confidence_from_risk_min and confidence_from_risk_max.
Number of SGM paths that give the same final disparity if SGM and its option activated, see Plugin libSGM. This band is named optimization_plugin_libsgm_nb_of_directions.
Left-right distance following cross checking method, see Validation of the disparity map. This band is named confidence_from_left_right_consistency.
Note
If more than one cost_volume_confidence is set on the pipeline (like for example cost_volume_confidence and cost_volume_confidence.after), then the corresponding band name will include the step indicator suffix (like .after in the example). For instance, with the following pipeline :
"cost_volume_confidence":
{
"confidence_method": "ambiguity",
"eta_max": 0.7,
"eta_step": 0.01
}
,
"cost_volume_confidence.after":
{
"confidence_method": "std_intensity"
}
The confidence bands will be named: confidence_from_ambiguity and confidence_from_intensity_std.after.
Note
Right products are only available if the validation step is present in the configuration file.
Validity mask
Validity mask indicates why a pixel in the image is invalid and provide information on the reliability of the match. These masks are 16-bit encoded: each bit represents a rejection / information criterion (= 1 if rejection / information, = 0 otherwise):
Bit |
Binary |
Description |
|---|---|---|
0 |
0000000000000001 |
The point is invalid, there are two possible cases:
- border of left image
- nodata of left image
|
1 |
0000000000000010 |
The point is invalid, there are two possible cases:
- Disparity range does not permit to find any point on the right image
- nodata of right image
|
2 |
0000000000000100 |
Information : disparity range cannot be used completely , reaching border of right image |
3 |
0000000000001000 |
Information : calculations stopped at the pixel stage, sub-pixel interpolation was not successful (for vfit, pixels d-1 and/or d+1 could not be calculated) |
4 |
0000000000010000 |
Information : filled occlusion |
5 |
0000000000100000 |
Information : filled mismatch |
6 |
0000000001000000 |
The point is invalid: invalidated by the validity mask associated to the left image |
7 |
0000000010000000 |
The point is invalid: right positions to be scanned invalidated by the mask of the right image |
8 |
0000000100000000 |
The point is invalid: point located in an occlusion area |
9 |
0000001000000000 |
The point is invalid: mismatch |
10 |
0000010000000000 |
Information : No data was filled |
11 |
0000100000000000 |
Information : Interval was in a regularization zone during filtering |
Memory consumption estimation
Pandora can give an estimation of the memory consumption of a given pipeline without running it.
The memory consumption estimation is obtained given the following graph, which shows the memory consumption of the
10 most consuming or used Pandora’s functions in respect of the size of the cost volume in MiB, defined as :
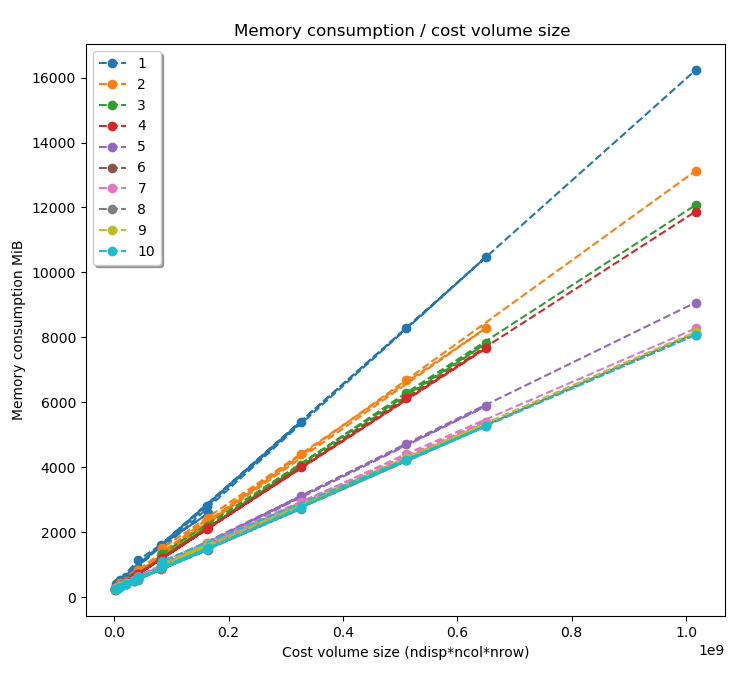
Being the functions :
Mc_cnn.run_mc_cnn_fast/accurate
Plugin_libsgm.optimize_cv
Aggregation.cost_volume_aggregation cbca
Matching_cost.compute_cost_volume sd/sad
Disparity.mask_invalid_variable_disparity_range/to_disp
Cost_volume_confidence.confidence_prediction ambiguity/std_intensity
Validation.Interpolated_disparity sgm
Matching_cost.compute_cost_volume census
Filter.bilateral_kernel
Matching_cost.compute_cost_volume zncc
The equation of each function’s consumption has been obtained and since the maximum memory consumption of a pipeline is defined by its most consuming function, Pandora just needs to find the first function in the list that is present in the pipeline to estimate the pipeline’s memory consumption.